Last updated 5/2022
MP4 | Video: h264, 1280×720 | Audio: AAC, 44.1 KHz
Language: English | Size: 1.15 GB | Duration: 1h 25m
Autodock Vina Tutorial.
What you’ll learn
You will learn Drug Designing and to perform Molecular Docking.
You will learn about the Drugs which are based on the Protein -Ligand Interaction.
You will learn to perform Molecular Docking using Autodock Vina.
You will learn the analysis of Docking Result and visualisation on Protein-Ligand Complex.
Requirements
Basis knowledge of Biology and Computer Science is required.
Description
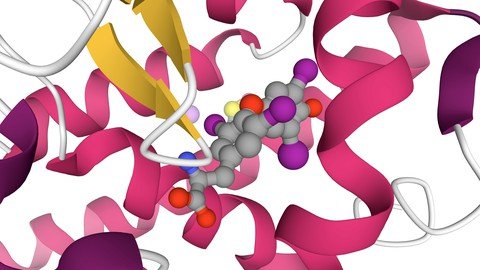
Molecular docking is a computational procedure that attempts to predict non-covalent binding of macromolecules or, more frequently, of a macromolecule (receptor) and a small molecule (ligand) efficiently, starting with their unbound structures, structures obtained from MD simulations, or homology modelling, etc. The goal is to predict the bound conformations and the binding affinity.The prediction of binding of small molecules to proteins is of particular practical importance because it is used to screen virtual libraries of drug-like molecules in order to obtain leads for further drug development. Docking can also be used to try to predict the bound conformation of known binders, when the experimental holo structures are unavailable.1One is interested in maximising the accuracy of these predictions while minimising the computer time they take, since the computational resources spent on docking are considerable.Molecular docking has become an increasingly important tool for drug discovery. In this review, we present a brief introduction of the available molecular docking methods, and their development and applications in drug discovery. The relevant basic theories, including sampling algorithms and scoring functions, are summarised. The differences in and performance of available docking software are also discussed. Flexible receptor molecular docking approaches, especially those including backbone flexibility in receptors, are a challenge for available docking methods. A recently developed Local Move Monte Carlo (LMMC) based approach is introduced as a potential solution to flexible receptor docking problems. Three application examples of molecular docking approaches for drug discovery are provided.Autodock Vina, a new program for molecular docking and virtual screening, is presented. AutoDock Vina achieves an approximately two orders of magnitude speed-up compared to the molecular docking software previously developed in our lab (AutoDock 4), while also significantly improving the accuracy of the binding mode predictions, judging by our tests on the training set used in AutoDock 4 development. Further speed-up is achieved from parallelism, by using multithreading on multi-core machines. AutoDock Vina automatically calculates the grid maps and clusters the results in a way transparent to the user.
Overview
Section 1: Introduction
Lecture 1 Introduction
Lecture 2 Schematic Illustration of Molecular Docking.
Lecture 3 Examples of Drugs based on Protein-Ligand Interaction.
Lecture 4 Autodock Vina: A Brief Introduction
Section 2: Performing Molecular Docking.
Lecture 5 Autodock Vina and MGL Tools Installation.
Lecture 6 Downloading Protein and Ligand for Docking.
Lecture 7 Treatment of Protein before docking and Grid Box Selection.
Lecture 8 Treatment of Ligand before Docking.
Lecture 9 Configuration File Setup.
Lecture 10 Run Autodock Vina to start Docking.
Lecture 11 Docking Result Analysis and Visualisation of protein ligand complex using PyMol.
Section 3: Assignments
Drug Designers , Bioinformaticians, Biochemists, Biologists.
Homepage
https://www.udemy.com/course/learn-molecular-docking-with-autodock-vina/
DOWNLOAD FROM RAPIDGATOR.NET
DOWNLOAD FROM RAPIDGATOR.NET
DOWNLOAD FROM UPLOADGIG.COM
DOWNLOAD FROM UPLOADGIG.COM
DOWNLOAD FROM NITROFLARE.COM
DOWNLOAD FROM NITROFLARE.COM



